|
We develop and apply computational structural modeling and design algorithms to predict and engineer proteins and their interactions, with a particular interest in immune recognition and therapeutics. Our work is highly interdisciplinary and we often work with experimental collaborators. Areas of particular interest, and selected highlights from that work, are given below.
|
T cell receptor modeling and design
T cell receptors (TCRs) are a key element of the adaptive immune system, with astounding diversity and exquisite specificity. We are developing algorithms to predict TCR structures from sequence, dock TCRs to peptide-MHC complexes, and to design TCRs to make improved immunotherapeutics. Our work in this area has led to the algorithms TCRFlexDock (TCR-peptide-MHC docking), ZAFFI (TCR design), and TCRmodel, a TCR structure modeling web server.
|
TCRFlexDock, Protein Science 2013
|
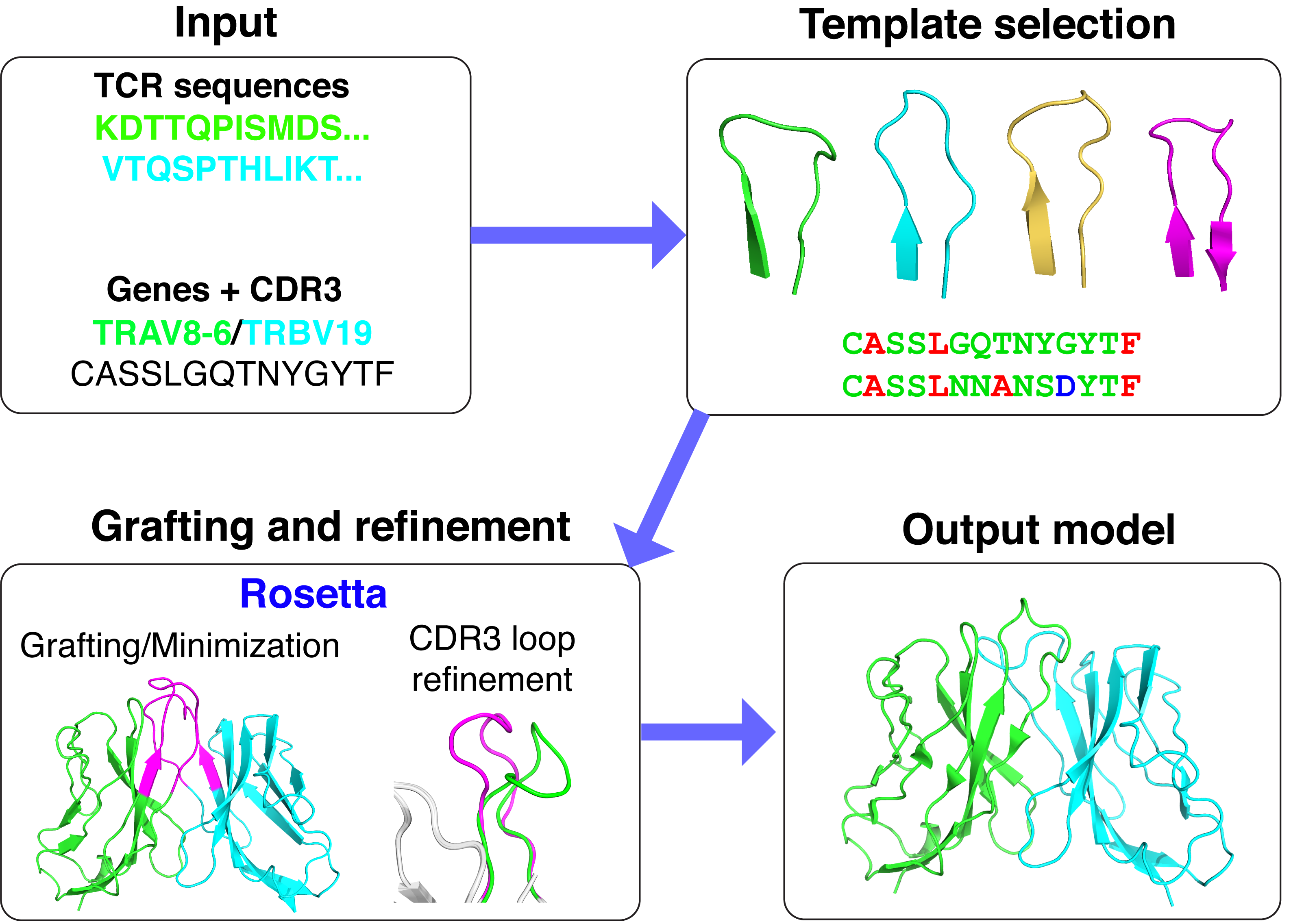
TCRmodel, Nucleic Acids Research 2018
|
|
Structure-based vaccine design
Many viruses and pathogens have no effective vaccines, however numerous advances and findings from immunology, high throughput screening, and structural biology provide the opportunity to use rational design approaches in this context. We are utilizing these advances and structural information to design new and effective vaccines for challenging targets, including hepatitis C virus (HCV). Related interests and research in the lab also include gaining insights into viral fitness, viral escape and antibody recognition through bioinformatics and structural modeling.
|
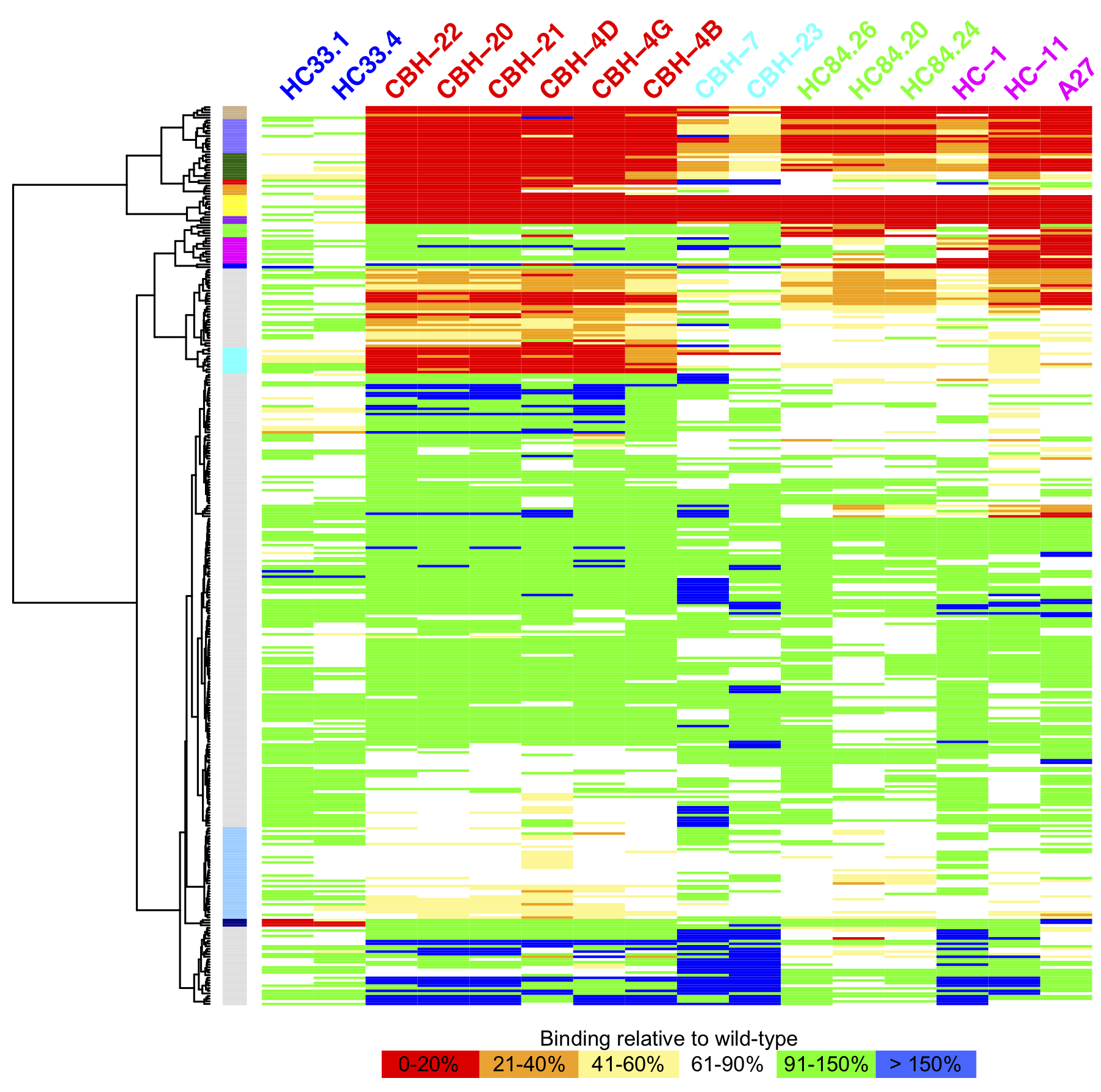
Global HCV E2 antibody mapping, PNAS 2016
|
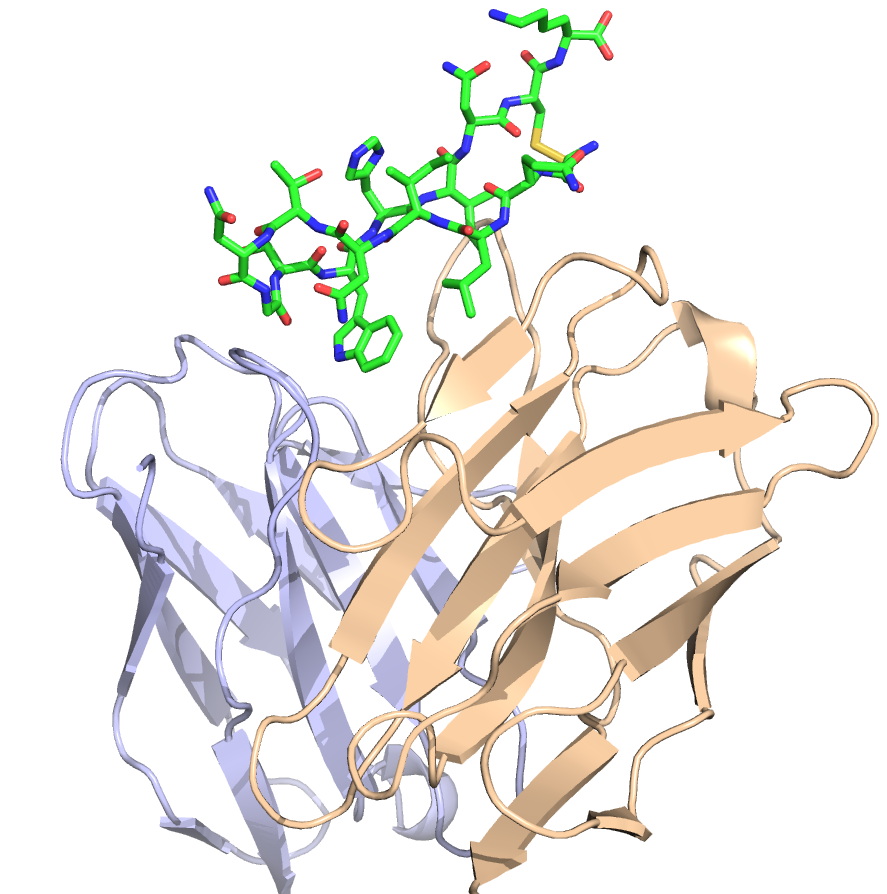
HCV vaccine design, Journal of Virology 2017
|